
This blog is a tutorial on some of the tools and features of HDF5 files. More specifically, we will cover the built-in terminal commands and some use of h5py, a HDF5 python package. You can download a VirtualBox image with required materials for the exercises from the following link (the username and password are both feeg6003).
Also, you can find the presentation slides here.
If you do not currently have access to HDF5 you can download it for free using macports or from their website. Acquiring the h5py module is very straightforward if you have an anaconda distribution installed, and we would recommend installing it using this method.
$ conda install h5py
Note that this installs the h5py module and the HDF5 tools at the same time.
What is HDF5?
HDF5 is a hierarchical file format that allows various types of data and metadata to be stored in the same file. It has a huge amount of structural versatility and can even be used for parallel applications.
The three main sections of an HDF5 file are groups, datasets and attributes. Groups can contain other groups and datasets and are used in a similar way to folders in a directory structure. Data is stored in datasets. Attributes contain the metadata and can be attached to either groups or datasets.
Part 1: HDF5 Command Line Tools
The HDF5 package comes with some helpful command line tools for use in viewing the files in the terminal. The first of these commands that we will mention is h5ls, which is a method very similar to the ls which is used in a terminal to look at the contents of a directory. The flags for this command are shown below.
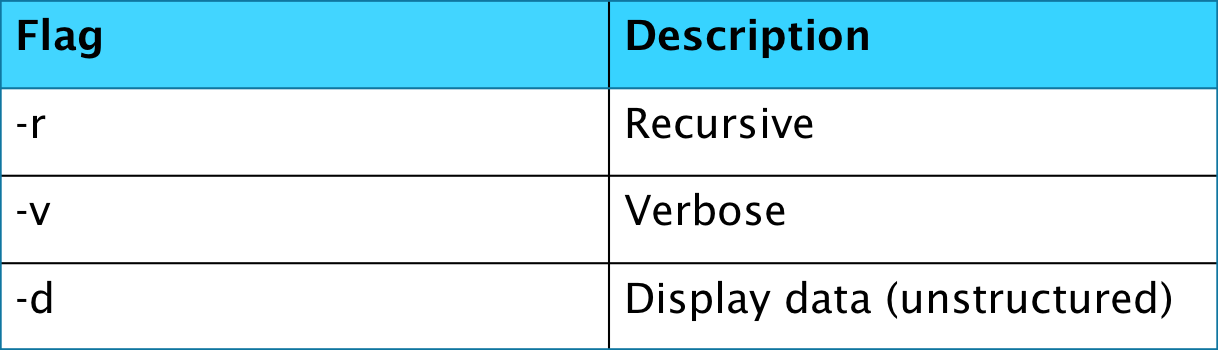
In addition to a filename, you may also provide h5ls with a path inside the file i.e.
$ h5ls file.h5/path
The second tool we will explain a bit about is the h5dump command. This is also a viewing tool but without using any flags is quite different to the aforementioned h5ls. It is a lot more verbose in its standard output, providing details of all data and even attributes and it is therefore recommended to initially surpress some of these details using flags if observing large HDF5 files.
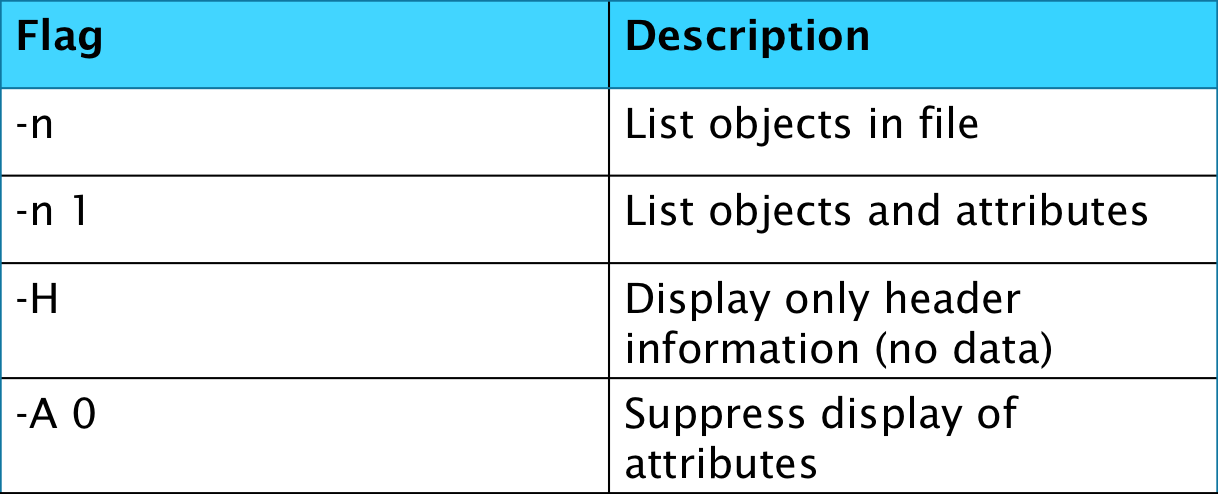
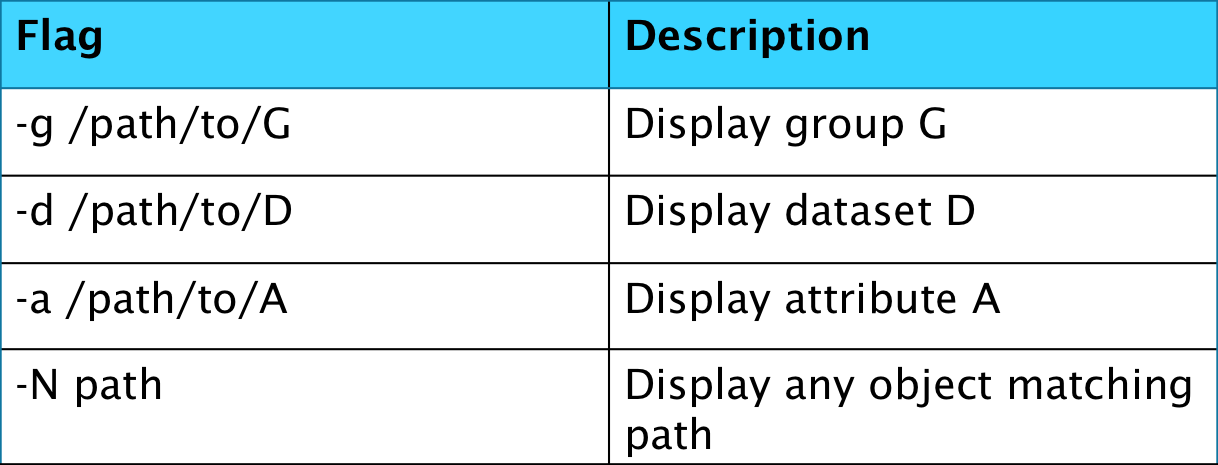
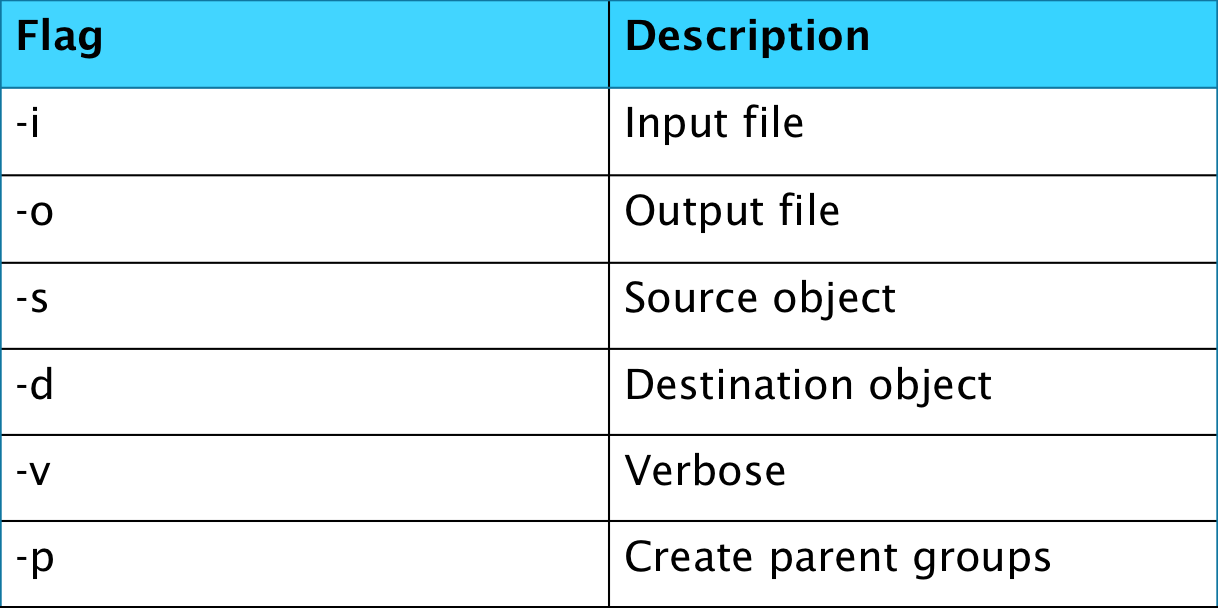
The final command line tool that will be covered in this blog is h5copy, which allows a user to copy datasets from one file to another or within a file.
Exercise 1: Find the Code
The first exercise for this tutorial will be based on using the three terminal comamnds discussed in the first section of the presentation; h5ls, h5dump and h5copy. In /Documents/ inside the VirtualBox image, you will find two HDF5 files named exercise1.h5 and solution.h5. The objective here is to search exercise1.h5, following clues in attributes to copy datasets to solution.h5. This exercise begins with a clue in the attribute of the root group. Keep on collecting the correct datasets and copy them over to another HDF5 named solution.h5 in the format specified by the attributes. When you have completed this task, run the python file "codebreaker.py" and it will check your solution.h5 to see if what you have submitted is correct.
Part 2: HDF5 and h5py
For those that are familiar with python, there is a module that provides various commands for the creation and manipulation of HDF5 files. Extensive documentation for this module can be found on this website.
This module has various features, including creation, reading and writing and the manipulation of file structure within HDF5 files. When wishing to use this module do not forget to use the "import h5py" command.
See the slides for details on the covered commands.
Exercise 2: Create your own file!
In the second part, you will have learned about using h5py and some of its features. This exercise will be about creating your own HDF5 file. In /Documents/ inside the VirtualBox image, you will find an IPython Notebook. Open this notebook through terminal using the command
$ cd ~/Documents/
$ ipython notebook exercise2.ipynb
Run through the exercises found in this file.